Closing the genomic data diversity gap: lessons from South Asia
The GenomeIndia Project is working to close the genomic data diversity gap and has opened its genetic database to researchers worldwide
8 May 2025
Keen to foster global collaboration, the GenomeIndia Project has just released its preliminary findings and opened its genetic database to researchers worldwide. This marks a critical step toward building a truly inclusive future for patient outcomes driven by precision medicine —one that reflects the complexity of human genetic diversity more accurately.
Precision medicine promises to transform healthcare by tailoring prevention, diagnosis, and treatment using genetic data. However, over 80% of the genomic data driving these advances comes from people of European ancestry, creating a biased foundation for medical research and clinical decision-making. The UK is advancing diverse genomic research through projects like Our Future Health and Genes & Health, which focus on sub-populations—but current categories remain too broad to fully capture genetic diversity. As the global conversation around equitable healthcare intensifies, GenomeIndia offers a practical model for how we could get diverse data representation right.
The GenomeIndia Project: a new model for inclusive genomics
The GenomeIndia Project, launched in 2020, represents a strategic effort to map India’s vast genetic diversity. It focuses on India’s rich tapestry of ethnolinguistic communities (Indo-European, Dravidian, Austro-Asiatic, and Tibeto-Burman). The approach is both academic and deeply practical. By identifying community-specific genetic risks, GenomeIndia enables more accurate diagnostics, personalised treatments, and affordable screening tools for conditions such as heart disease, diabetes, and rare genetic disorders.
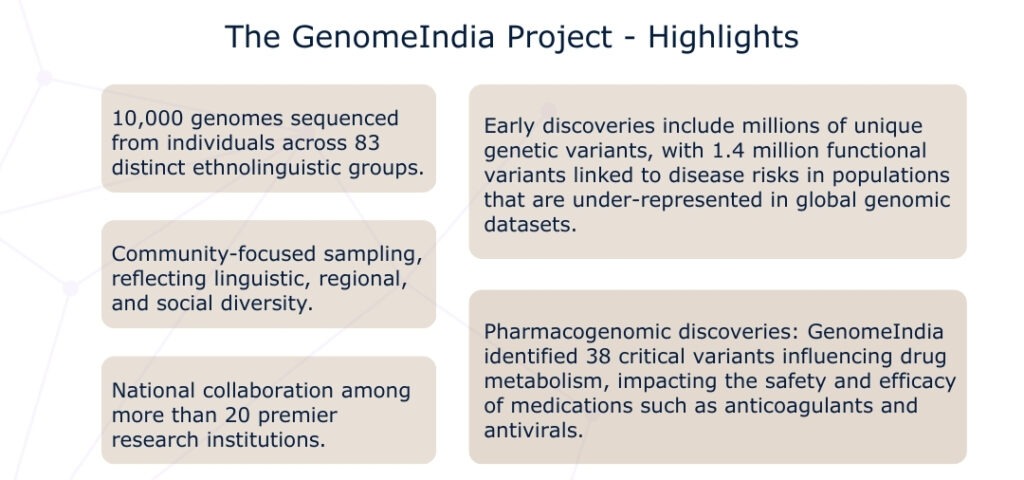
Why broad ethnic labels hold us back
The term “South Asian” is often used as a catch-all to group people from countries including India, Pakistan, Bangladesh, Nepal, Sri Lanka, Bhutan, and the Maldives (and sometimes Afghanistan). Yet in UK clinical settings, this vast and complex region is usually reduced to a few broad checkboxes such as “British Indian,” “British Pakistani,” or “Other South Asian.”
This oversimplification obscures the deep internal diversity of South Asian populations. For example, while South Asians are frequently reported to be at higher risk for conditions like Type 2 diabetes (T2DM), cardiovascular disease (CVD), and blood disorders (anaemias) these risks vary widely across subgroups, —such as Sri Lankans, Bangladeshis, and Punjabi-speaking populations from both India and Pakistan. Similarly, genetic traits like G6PD deficiency, which can lead to a type of anaemia, are concentrated in specific ethnic subgroups in Pakistan and Afghanistan—not uniformly across “South Asians”.
Compounding the issue is the lack of a standardised system for recording ethnicity in healthcare. Ethnic data are often recorded inconsistently across health systems, making it difficult to identify population-specific risks. Reports from the NHS Race and Health Observatory show that this misclassification contributes to critical gaps in both research and healthcare delivery.
Efforts like those by the National Academy of Sciences, Engineering and Medicine are underway to improve the categorisation of populations in genomic studies. However, much more work is needed to account for evolutionary history, cultural practices, and environmental contexts – all of which shape human genetic variation far more accurately than racial or national labels.
Why evolutionary ancestry and ethnolinguistic identity matter
Human genetic diversity does not conform to modern political borders or racial categories. Instead, it reflects thousands of years of migration, adaptation, isolation, and cultural practices like endogamy (marrying within one’s own social or cultural group). One of the most powerful ways to understand this genetic variation is through ethnolinguistic identity – the deep-rooted community bonds shaped by language, geography, and cultural lineage.
South Asia offers a vivid case study. The region is home to more than 4,500 anthropologically recognised groups, many of which have retained genetic distinctiveness for generations due to strict patterns of endogamy. Ignoring this complexity doesn’t just miss important medical insights—it also perpetuates inequities in healthcare access and outcomes.
Yet, despite comprising nearly 25% of the world’s population, South Asians make up less than 2% of participants in global genome-wide association studies (GWAS). This glaring underrepresentation leads to major blind spots in research—especially in areas like drug metabolism, disease risk, and treatment efficacy.
For instance, a genomic study involving over 1,000 individuals across diverse Indian subpopulations found significant variation in drug-related genes such as CYP2C9, CYP2C19, and NAT2. These genes influence how people process medications like warfarin, commonly prescribed for heart conditions. Variants in these genes didn’t just differ from European populations—they varied widely within South Asian subgroups. This highlights the limits of using umbrella categories like “South Asian” in fields like pharmacogenomics.
Why this matters globally
The challenges of underrepresentation and oversimplified categorisation aren’t unique to South Asia. The historical forces that shaped its genomic diversity—migration, isolation, linguistic shifts—are mirrored in other parts of the world, including Africa, Southeast Asia, and the Americas.
To ensure equitable healthcare worldwide, we need tailored data collection methods that respect local cultural practices and ethical considerations. Many of these regions have histories of exploitation and mistrust in research, making community-driven and ethical frameworks even more critical.
The GenomeIndia Project is an example of how to do this responsibly. Its use of the Biotech-PRIDE guidelines ensures that data is shared ethically, participants are protected, and local interests are prioritised—setting a powerful blueprint for inclusive genomics worldwide.
Why underrepresentation affects everyone – and how the UK can lead
The UK, with its large South Asian diaspora, is well-positioned to lead in using this newly released GenomeIndia diverse data source. Revamping existing health data to make it more representative would be costly and complex, but partnering with GenomeIndia offers a more practical solution.
By leveraging the UK’s vast sequencing expertise and datasets, and collaborating with the GenomeIndia team, they can conduct clinically relevant studies to identify genetic risk factors and develop low-cost diagnostic tools which are more comprehensive and sensitive to diverse genetic variants from non-European populations as well. These tools, designed to improve early detection of conditions like diabetes and heart disease, would not only enhance global health datasets but also improve care in the UK.
By adopting affordable and suitable diagnostic solutions, the UK can reduce the financial burden on the NHS while providing more precise and cost-effective care. This partnership can set a new standard for inclusive, equitable healthcare innovations, benefiting healthcare worldwide and grounding advancements in global genetic diversity.
A blueprint for the future: global genomic inclusion
GenomeIndia isn’t alone. Global efforts to close the genomic diversity gap are underway, with initiatives like 3MAG in Africa mapping African genetic variation, and Origen in Mexico working to include indigenous communities in genomic studies.
The future of precision medicine and its potential to improve patient’s lives hinges on who is represented in our data. Projects like GenomeIndia are charting a path forward—one that is scientifically sound, ethically responsible, and globally inclusive.
By embracing the evolutionary, linguistic, and cultural complexity of human populations, we can build systems of care that are more effective, equitable, and just to individuals. By supporting these efforts—whether through collaboration, policy change, or public awareness—countries with vast genomic expertise like the UK can play a vital role in shaping that future.

- DNA computers: invisible monitors of health?
- First do no harm: shaping health AI governance in a changing global landscape
- Delivering diagnosis for prenatal medicine – lessons from implementation of a national service
- Metagenomic sequencing in public health – can long read sequencing be used to tackle antimicrobial resistance?
